This article is more than 1 year old
Online gamers strike major blow in battle against AIDS
Distributed human-brain cluster cracks protein conundrum
A bunch of gamers have untangled the structure of a key protein in the virus that causes AIDS, a mystery that has left scientists stumped for decades.
It took just three weeks for players of online game Foldit to predict an accurate model for the protein – a type of enzyme called a retroviral protease that has a critical role in how the HI virus grows and spreads.
The game Foldit was specially designed to help work out the structure of proteins. It combines computer intelligence with human spatial abilities by asking players to tweak and tug 3D models, a task that computers find hard to do.
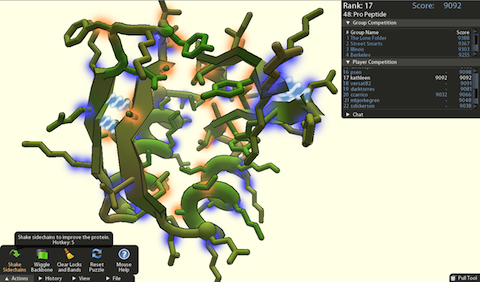
A screengrab of online protein-folding game, Foldit
The gamers – most of whom have no background in biochemistry – generated models good enough for the researchers to refine and, within a few days, determine the enzyme's structure. In the research published yesterday in Nature Structural & Molecular Biology, lead authors biochemists Firas Khatib and Frank DiMaio gave gamers equal billing with the rest of their biochemist co-authors.
Amazingly, surfaces on the molecule stood out as likely targets for drugs to de-active the enzyme. "These features provide exciting opportunities for the design of retroviral drugs, including AIDS drugs," wrote the authors of the paper.
The solution of the virus enzyme structure, the researchers said, "indicates the power of online computer games to channel human intuition and three-dimensional pattern matching skills to solve challenging scientific problems."
Proteins are made of long chains of amino acids that tend to fold up in particular ways. Understanding the shapes that the amino acids fold up into is important in working out what they do, and – in the case of disease proteins – finding means of stopping them doing it. The scientists behind Foldit explain:
Every kind of protein folds up into a very specific shape – the same shape every time [...] The more we know about how certain proteins fold, the better new proteins we can design to combat the disease-related proteins and cure the diseases.
The research was published on 18 September in Nature Structural & Molecular Biology.
A PDF of the research is available here (3-page/812KB PDF)
